RXBio Translates Sequence to Science and Industry
Tel: 027-87050299Email: sales@rxbio.cc
- Home
- Single-cell Sequencing
- Spatial Transcriptomics
- Third-generation Sequencing
- Omics Technologies
- Bioinformatics
- Experimental Platform
- About US
中文
RXBio Translates Sequence to Science and Industry
The Stereo-CITE Proteo-Transcriptomics Set enables simultaneous in situ detection of the whole transcriptome and 100+ proteins on the same tissue section — all with subcellular resolution. By integrating high-plex protein profiling with unbiased transcriptomics in a single experiment, Stereo-CITE empowers researchers to decode cellular phenotypes, interactions, and tissue architecture with unprecedented depth and clarity.
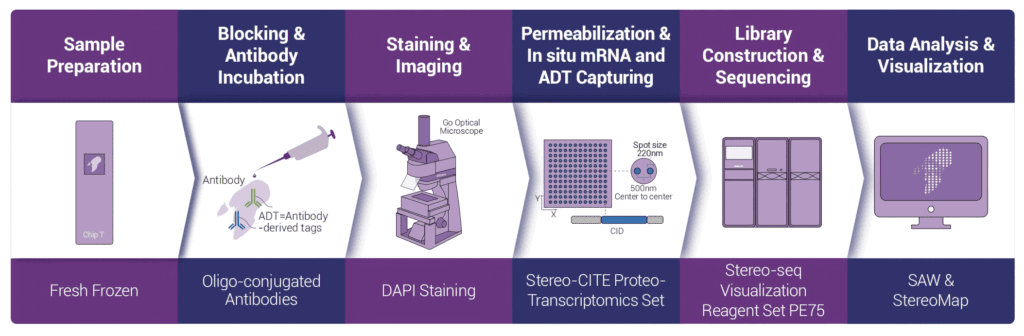
Utilizing the ultra-high resolution of Stereo-seq, Stereo-CITE combines Cellular Indexing of Transcriptomes and
Epitopes by Sequencing (CITE-seq) with the Stereo-seq workflow. It captures mRNA and antibody-derived tags (ADTs) at nanoscale resolution across a centimeter-scale field of view. Through a streamlined “tissue-to-data” workflow, the system delivers spatial maps of transcriptome and protein expression via sequencing and visualization.
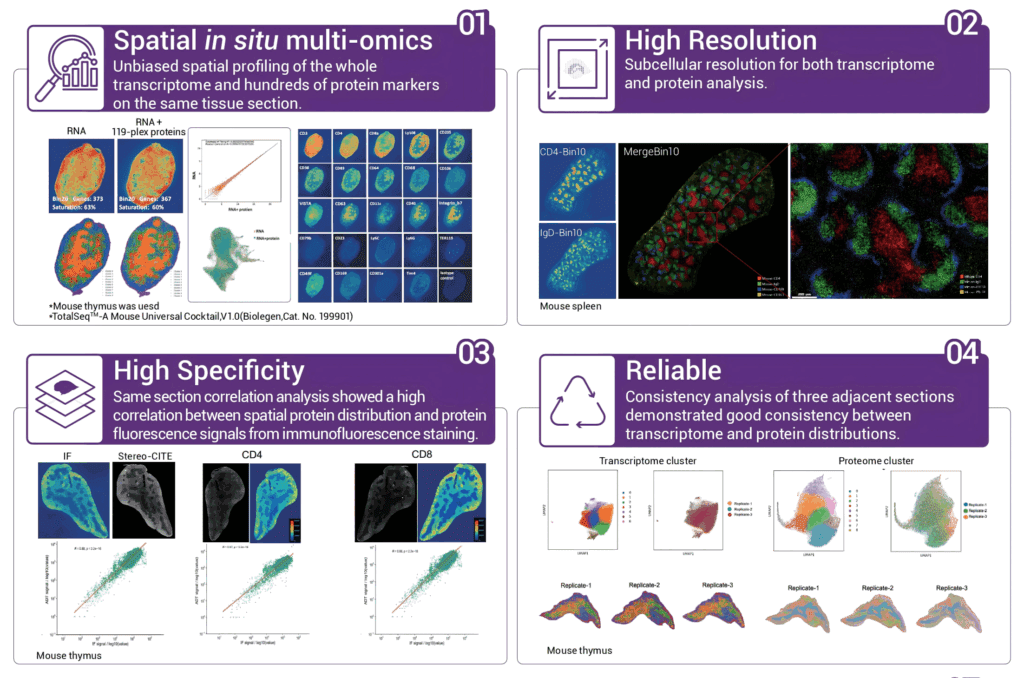
1. Unbiased spatial in situ multi-omics
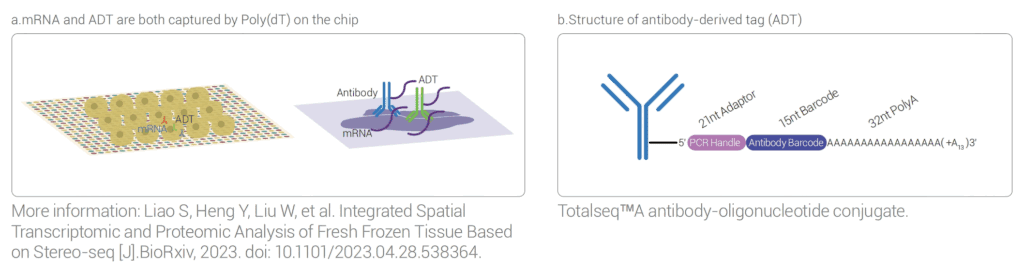
It can achieve unbiased co-detection of RNA and proteins on the same tissue slice; protein detection does not affect the capture of transcriptome genes.
2. Ultra-high multiplex protein detection.
Efficiently detect as many as 100+ multiplex proteins and support the free combination of antibodies.
3. Single-cell or subcellular level resolution.
With a resolution of 500 nm, it presents the molecular landscape at the spatial micro-scale.
4. Sequencing-based spatial protein detection.
There is no interference from autofluorescence; there is no antigen instability caused by multiple rounds of cyclic detection; the increase in the multiplicity of protein detection does not affect the detection duration.
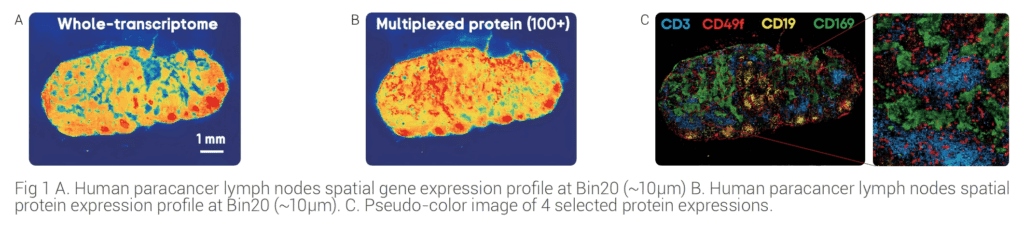
Applications of Stereo-CITE in the multi-omics study of human paracancer lymph nodes
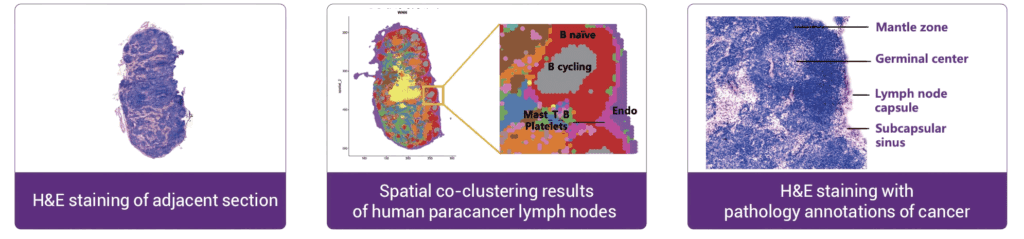
Stereo-CITE recapitulated the organizational structure of human lymph nodes
The figure below shows the comparison of the spatial co-clustering results with the H&E-stained image of adjacent
sections. It was found that the multi-omics spatial co-clustering results perfectly recapitulated the tissue structure of
the lymph nodes, which is highly consistent with prior biological and histological knowledge.
The figure below shows the comparison of the spatial co-clustering results with the H&E-stained image of adjacent
sections. It was found that the multi-omics spatial co-clustering results perfectly recapitulated the tissue structure of
the lymph nodes, which is highly consistent with prior biological and histological knowledge.
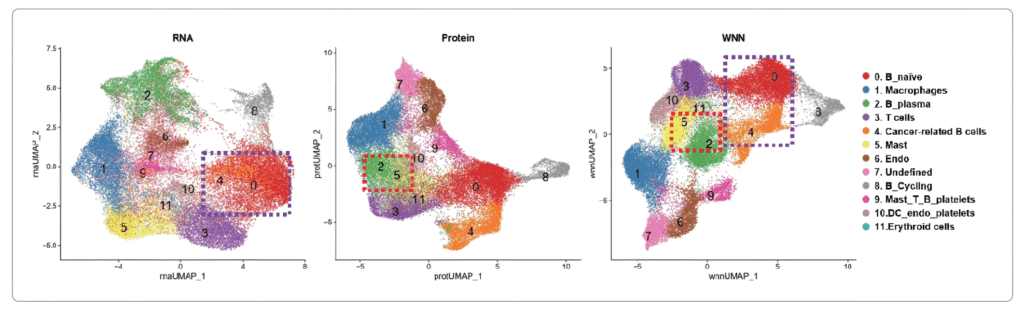
Joint analysis of RNA and protein information provides more accurate spatial clustering
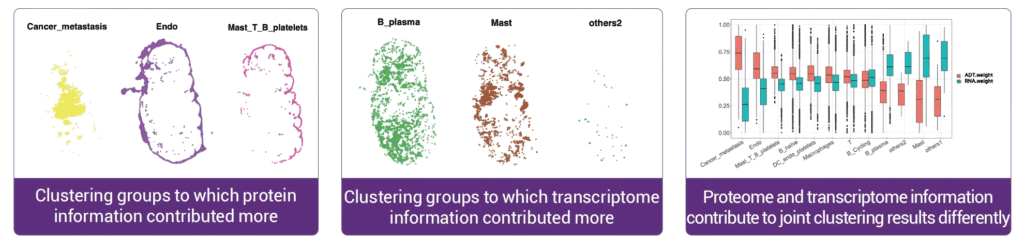
Protein and transcriptional information contribute to the clustering results differently. Statistical diagram
demonstrated that proteomic information plays a key role in grouping cancer metastatic related cells, endothelial cells,
mast cells and others, where transcriptome information is the main contribution in the cell type identification of
B_plasma and Mast groups.