introduction
In 1958, Crick put forward the “Central Dogma”, which outlined the information transfer among DNA, RNA, and proteins. Genes are transcribed into precursor RNA, and through alternative splicing, different transcripts are formed and then translated into protein isoforms with different functions. Alternative splicing allows a gene to produce multiple protein isoforms, achieving specific expression. Recently, “Nature Reviews Drug Discovery” has discussed the importance of protein isoforms in drug discovery and suggested using these isoforms to improve drug specificity and efficacy. Therefore, studying the regulatory mechanisms of alternative splicing of precursor RNA is crucial for the development of treatment methods based on protein isoforms.
RNA modifications are key regulators in cell biology, influencing the generation, transport, function, and metabolism of RNA. The major RNA modifications include m6A, m6Am, m1A, m5C, ac4C, m7G, Ψ, and A-to-I editing. These modifications are added, removed, recognized, and edited by specific RNA-modifying enzymes, thereby regulating cellular functions.
Among the second-generation NGS technologies, short-read RNA-seq is widely used to analyze gene expression and alternative splicing, often combined with polyA enrichment or rRNA depletion. PAS-seq focuses on locating the polyadenylation sites of mRNA, while PolyA-seq analyzes the changes in the length of the polyA tail, which is important for understanding mRNA stability and cell fate. Research on RNA modifications is also crucial as it affects RNA stability and function. Multiple methods such as IP capture and chemical conversion have been developed to reveal the diversity of RNA modifications. To comprehensively study gene expression and modifications, it is necessary to use multiple NGS technologies in combination.
Oxford Nanopore’s third-generation sequencing platform can directly sequence natural RNA strands.ONT-Direct RNA Sequencing (DRS) technology does not require the synthesis of a second cDNA strand and PCR amplification, avoiding the introduction of errors, directly obtaining mRNA sequences, and retaining information on RNA base modifications, accurately estimating the length of the poly(A) tail and revealing the true characteristics of RNA.
advantages
✔ Improve gene annotation: Discover new transcripts and genes, and obtain a comprehensive transcriptome map.
✔ Differential analysis: Differentially expressed genes (DEG), differentially expressed transcripts (DET), differentially used transcripts (DTU).
✔ Gene structure analysis: Accurately analyze alternative splicing (AS), polyadenylation sites (APA), the length of 3’UTR, and the length of the polyA tail.
✔ RNA modification analysis: m6A, m1A, m5C, m7G, hm5C, Ψ, etc.
✔ Transcript resolution: Analyze the regulatory mechanisms and functions of gene expression at the transcript level.
workflow
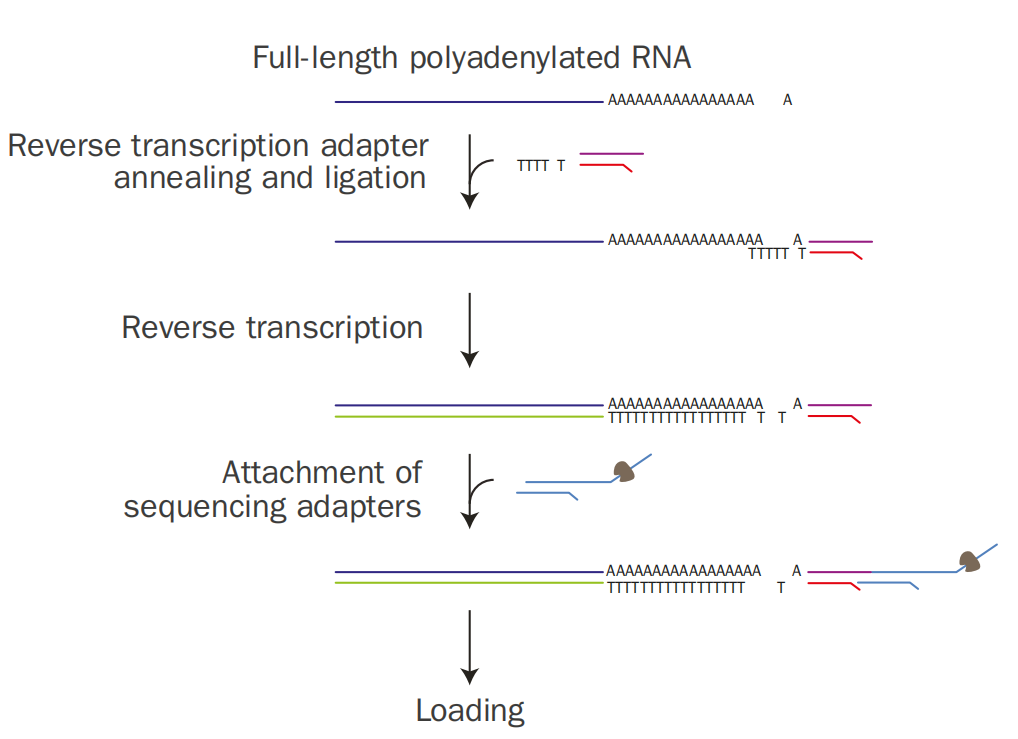
Firstly, the primer with an oligo(dT) end anneals and ligates to the PolyA tail of mRNA, and then reverse transcription is carried out to form an RNA-DNA hybrid strand. Subsequently, sequencing adapters linked with molecular motors are added for subsequent sequencing.
research Cases

The research group led by Chen Kaifu from Harvard Medical School/Boston Children’s Hospital and the research group led by Cao Qi from Northwestern University published a research paper titled “2′-O-methylation at internal sites on mRNA promotes mRNA stability” online in the journal “Molecular Cell”. A large number of studies have shown that stable mRNA is very important for the development of RNA vaccines and RNA therapies. In the latest research reported in this paper, Cao Qi et al. conducted a systematic analysis of the impact of Nm-modified mRNA on stability. The results showed that Nm-modified mRNA exhibited higher stability compared to unmodified mRNA. Compared with the control group, the mRNA bound by the FBL protein also showed enhanced stability. When the expression of FBL was inhibited, the stability and RNA levels of both Nm-modified mRNA and the mRNA that usually binds to FBL decreased significantly, indicating that the mRNA Nm modification catalyzed by FBL can effectively improve the stability of mRNA and thus increase the transcriptional level.
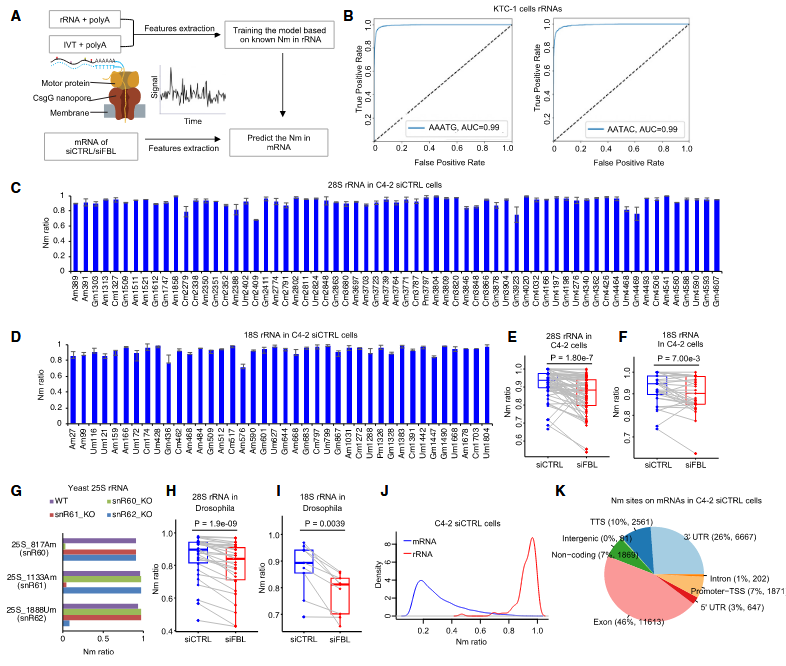
In this study, the authors innovatively combined the Oxford Nanopore direct RNA sequencing technology with machine learning algorithms and developed a new technique named NanoNm, which aims to identify Nm modifications. The practicability of this technique has been verified in ribosomal RNA samples from humans, yeast, and fruit flies. By applying the NanoNm technique, the researchers observed the phenomenon that the knockdown of the FBL gene led to a decrease in the level of Nm modifications in tumor-related mRNAs, thus revealing the prevalence of Nm modifications in mRNAs. Further analysis indicated that in the mRNA of PSMD13, a key driver gene for prostate cancer, the presence of Nm modifications was associated with higher stability. This finding reveals that the FBL gene promotes the malignant progression of prostate cancer by catalyzing the Nm modifications of oncogene mRNAs at the post-transcriptional level and enhancing their stability.
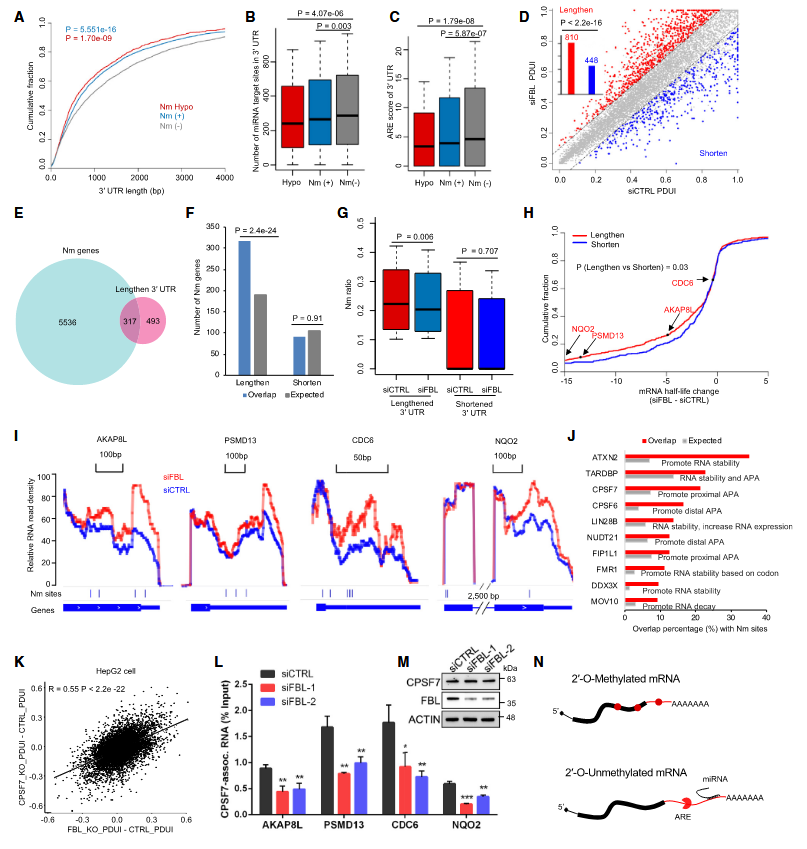
In the field of mechanism research, this study revealed that genes with Nm modifications tend to have shorter 3′ untranslated regions (3’UTR), and the downregulation of FBL can significantly affect the length of 3’UTR. This finding implies that FBL-mediated Nm modifications may enhance the stability of mRNA by regulating the variability of 3’UTR. Based on this, this study speculates that RNA-binding proteins may participate in the regulation of alternative polyadenylation (APA) process by recognizing Nm-modified mRNA and interacting with it. Through the integration of the analysis of the CLIPdb RNA-binding protein public database, the research results show that the binding sites of proteins such as ATX2 and CPSF7 highly overlap with Nm modification sites, which suggests that these proteins may be responsible for recognizing and binding to Nm-modified mRNA. Meanwhile, this study found that RNA-binding proteins, such as CPSF7 which is known to be involved in RNA stability or the regulation of the APA process, may affect the stability of mRNA by specifically recognizing Nm modifications. In the case of simultaneous knockout of FBL and CPSF7, the impact on the APA pattern was observed to be consistent with that of the single knockout of FBL, indicating that CPSF7 can indeed recognize Nm modifications and plays a crucial role in the regulation of RNA stability mediated by Nm modifications.
Reference
【1】 Kjer-Hansen, P., Phan, T.G. & Weatheritt, R.J. Protein isoform-centric therapeutics: expanding targets and increasing specificity. Nat Rev Drug Discov (2024).
【2】Li Y, Yi Y, Gao X, et al. 2′-O-methylation at internal sites on mRNA promotes mRNA stability. Molecular Cell. 2024 Jun;84(12):2320-2336.e6.
