RXBio Translates Sequence to Science and Industry
Tel: 027-87050299Email: sales@rxbio.cc
- Home
- Single-cell Sequencing
- Spatial Transcriptomics
- Third-generation Sequencing
- Omics Technologies
- Bioinformatics
- Experimental Platform
- About US
中文
RXBio Translates Sequence to Science and Industry
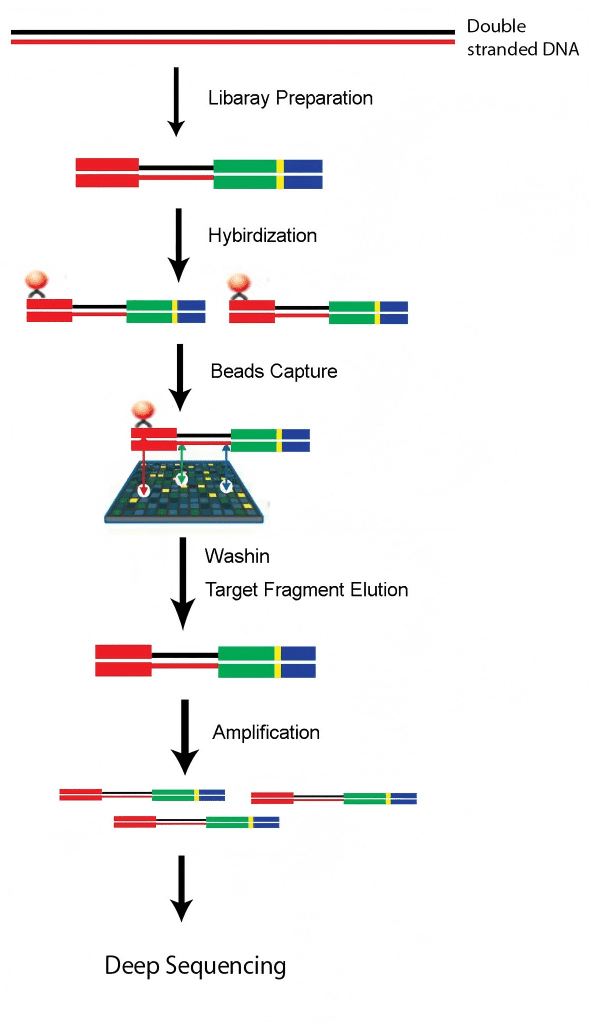
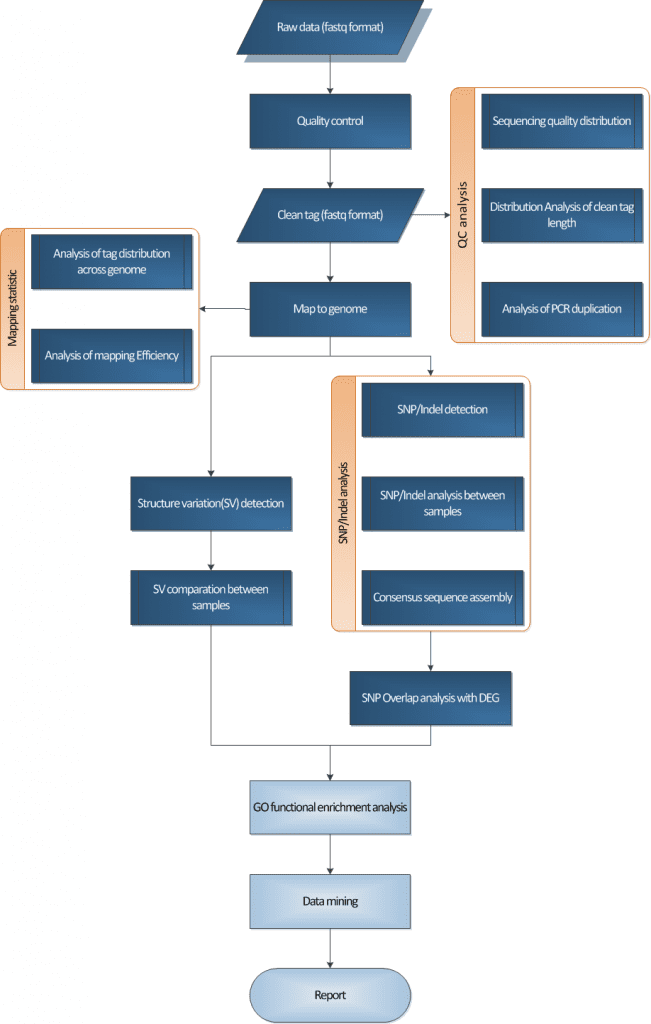
Whole exome sequencing (WES) is a genomic analysis method that uses sequence capture technology to capture and enrich the DNA in the exome regions of the whole genome and then conduct high-throughput sequencing. It can quickly detect variations in exons (coding regions) and splice sites. Compared with whole-genome sequencing, it is simpler, more economical and more efficient, and it has a higher coverage of the target regions, facilitating the detection of variations. Exonic mutations have been proven to be the cause of most monogenic diseases. In the years following the emergence of WES technology, the identification of genes for Mendelian diseases has increased significantly. Besides its application in the research of Mendelian diseases, currently, some complex diseases also use exome sequencing technology to discover relevant variations of diseases, such as cancer, cardiovascular diseases, Alzheimer’s disease, autoimmune diseases and so on.
✔ Provide personalized solutions for different research fields.
✔ The analysis process is comprehensive, with detailed data interpretation, and personalized analysis can be provided for later data mining.
✔ Utilize mainstream exome capture sequencing platforms, ensuring reliable data and good stability.
✔ Offer a variety of variation analyses: single nucleotide polymorphisms, insertions and deletions, nonsense mutations, structural variations, and copy number variations.

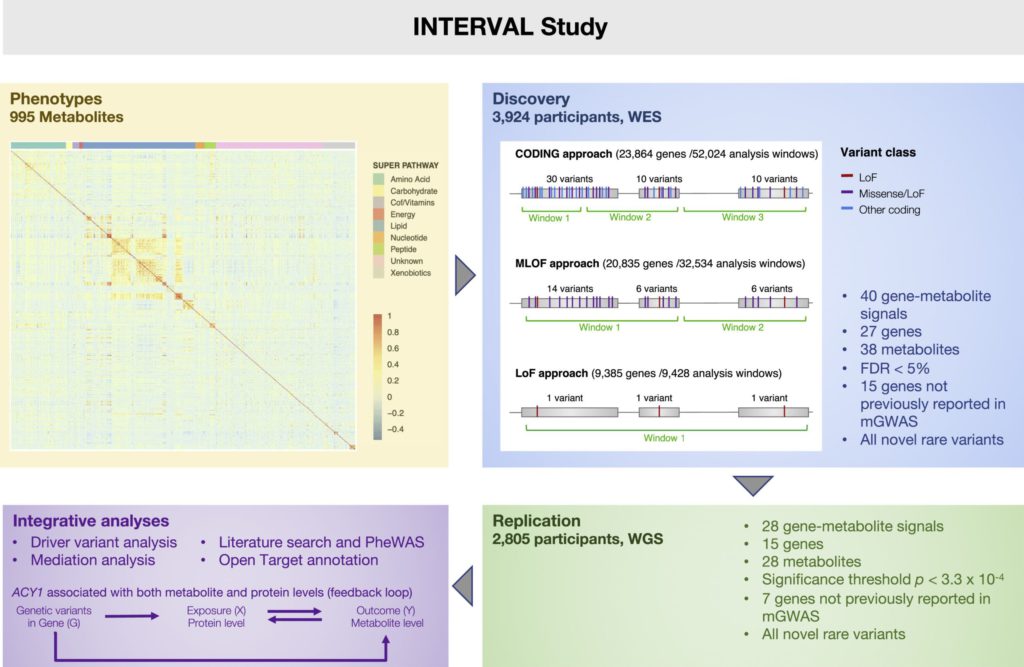
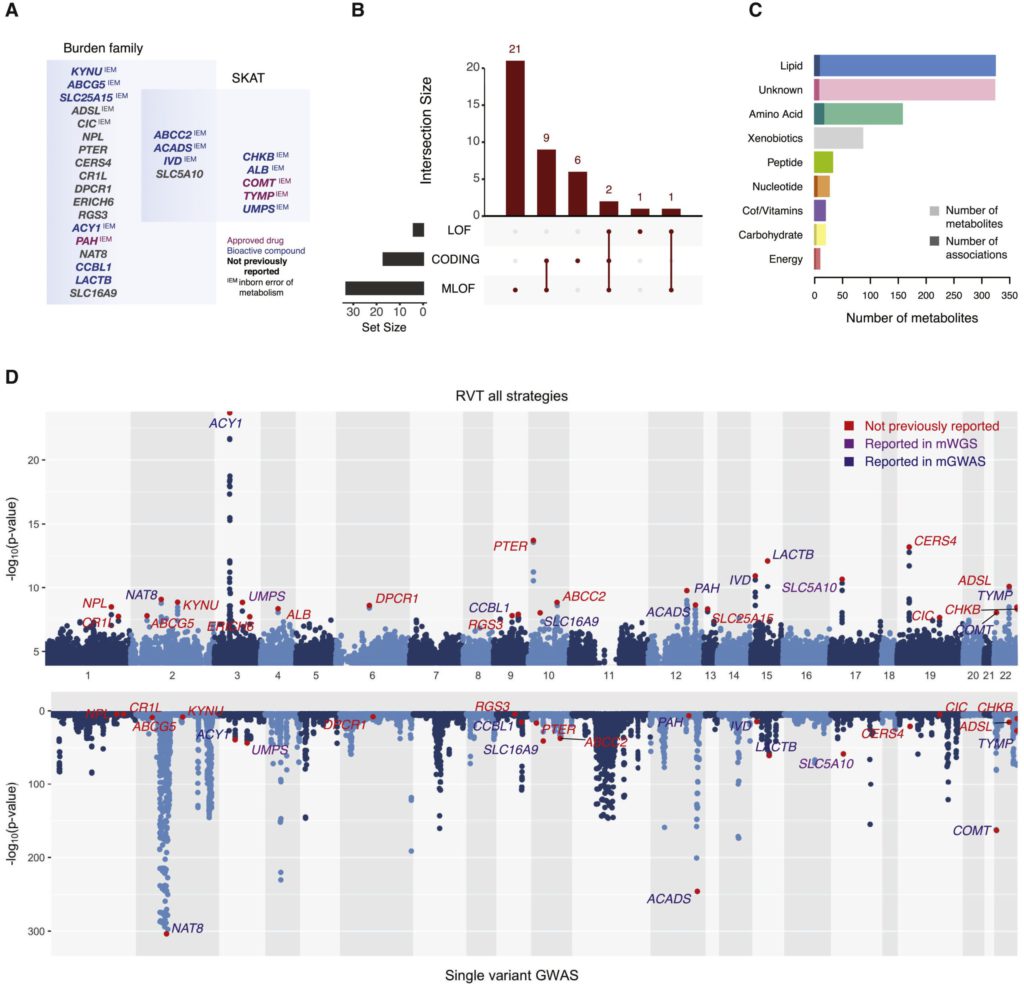
This study employed multiple methods to select and aggregate rare genetic variants in protein-coding regions and used ultra-high-performance liquid chromatography-tandem mass spectrometry to detect their correlations with 995 metabolites measured in plasma. Forty new associations involving rare coding variants (27 genes and 38 metabolites) were identified, among which 28 (15 genes and 28 metabolites) were replicated. An algorithm was developed to prioritize the putative driver variants at each locus, and mediation and Mendelian randomization analyses were used to test the directionality of the associations between metabolite and protein levels at the ACY1 locus. Overall, 66% of the reported associations involved gene targets of approved drugs or bioactive drug-like compounds, contributing to the efforts in drug target validation.