Introduction
An important feature of the activity of DNA regulatory elements lies in their chromatin accessibility, that is, the accessibility and operability of these elements within the chromatin structure. Only those active regulatory elements have the possibility of being recognized by the internal mechanisms of the cell and further expressed. Therefore, chromatin accessibility plays a crucial role in the process of transcriptional regulation. It provides key clues for studying how transcription factors influence gene expression by regulating chromatin accessibility.
In 2013, the research team led by Howard Y. Chang at Stanford University in the United States developed ATAC-seq, a method for studying chromatin accessibility. Its principle is based on the property that the transposase Tn5 can easily bind to open chromatin, and then the DNA sequences captured by the Tn5 enzyme are sequenced. Due to its requirement for a small amount of cells, simple experiment process, and the ability to detect chromatin accessibility across the whole genome, ATAC-seq has become the preferred technical method for related research.
advantages
✔ Low starting amount: The number of cells required ranges from 10,000 to 50,000 cells, and it can even be used for research at the single-cell level.
✔ Simple workflow: The steps are straightforward and the success rate is high.
✔ High specificity: It can effectively reduce the contamination of mitochondrial DNA data.
✔ Good reproducibility: The results of repeated experiments are highly consistent.
applications
1. Mapping the open chromatin landscape is of great significance for researching genomic structure and function, as well as disease diagnosis and treatment.
2. Identifying transcription factors involved in gene regulation through motif analysis is very important for understanding the complex mechanisms of gene expression and the occurrence mechanisms of gene-related diseases.
3. Identifying the target genes and functional elements regulated by transcription factors and studying the important roles of transcription factors in life activities such as cell growth, differentiation, and apoptosis are also important ways to understand the complex mechanisms of gene expression.
4. Combining nucleosome prediction analysis to accurately determine the positioning rules and influencing factors of nucleosomes can provide a more comprehensive understanding for the research on chromatin structure and function.
5. Obtaining the correlation information between gene expression and upstream regulatory elements and transcription factors through multi-omics combined analysis, and analyzing the variation mechanism of targeted regulation by transcription factors.
workflow

The core technical enzyme of ATAC-seq, the Tn5 transposase, can efficiently insert sequencing adapters into the accessible regions of chromatin. However, other regions of chromatin are tightly bound to histones and exist in the form of nucleosomes, forming a compact structure that restricts the insertion of the transposase. The Tn5 transposase can accurately insert the sequencing adapters directly into the double-stranded DNA in these open regions and fragment the DNA. Subsequently, these DNA fragments undergo subsequent processing such as extraction and PCR amplification, and finally, libraries suitable for next-generation sequencing are prepared. Through sequencing and data analysis, we can further obtain important information about open chromatin, which includes key biological information such as the binding sites of transcription factors (TF) and histone modification sites.
Demo Data
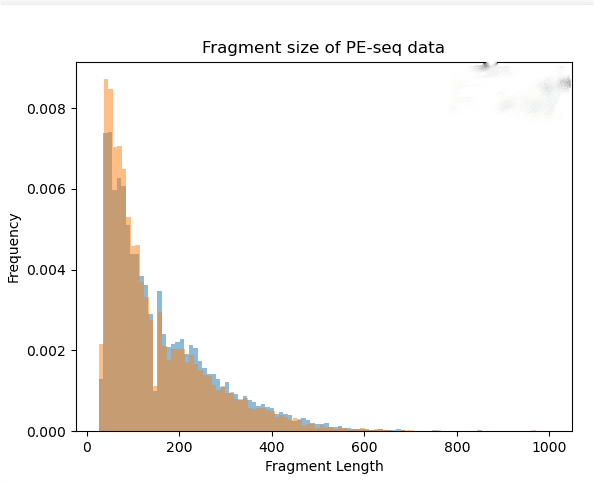
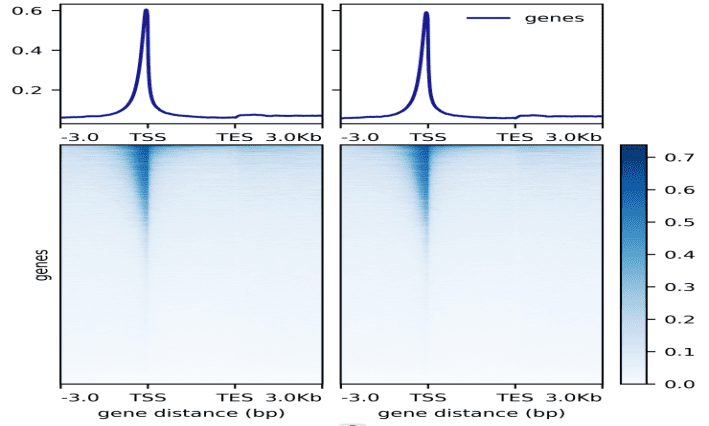
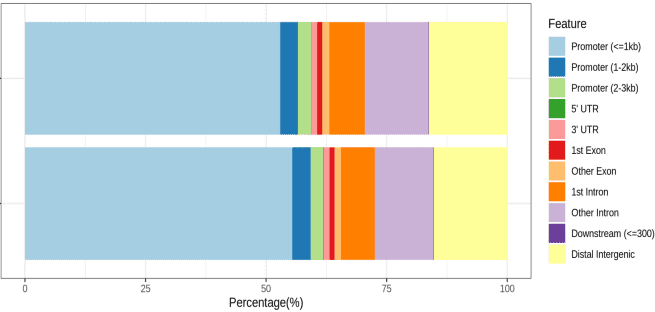
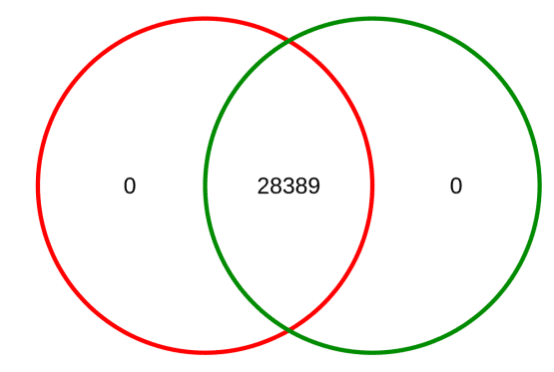
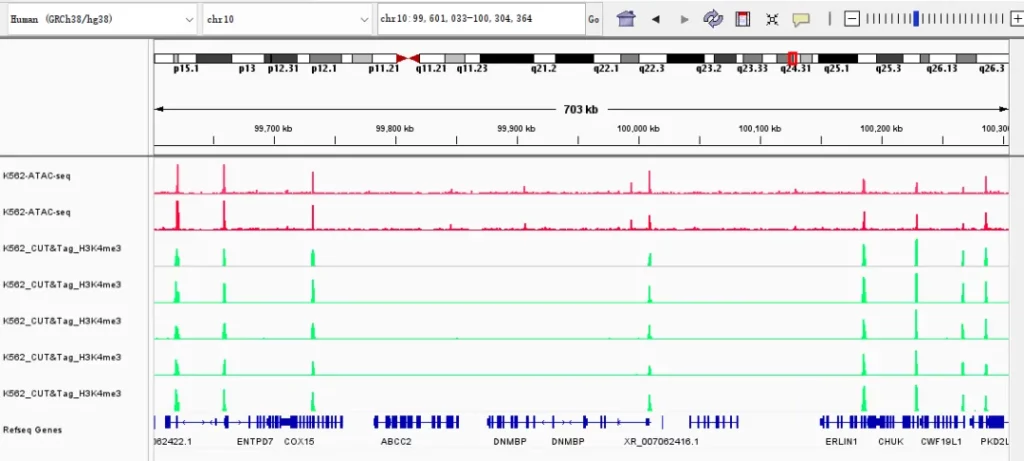
Research Cases
On October 26, 2018, the team led by Professor Howard Y. Chang published the latest achievements in chromatin accessibility research in the international top academic journal “Science”. Using the ATAC-seq method and combining with the existing data from The Cancer Genome Atlas (TCGA) research, the researchers located 562,709 reproducible and transposase-accessible chromatin accessibility sites in 410 frozen samples covering 23 cancer types. In the pan-cancer analysis of chromatin accessibility peaks, approximately two-thirds were consistent with the previously discovered regulatory element information, indicating that the ATAC-seq technology can well reproduce the findings of previous studies and can also discover a large number of new chromatin accessibility sensitive sites at the same time.
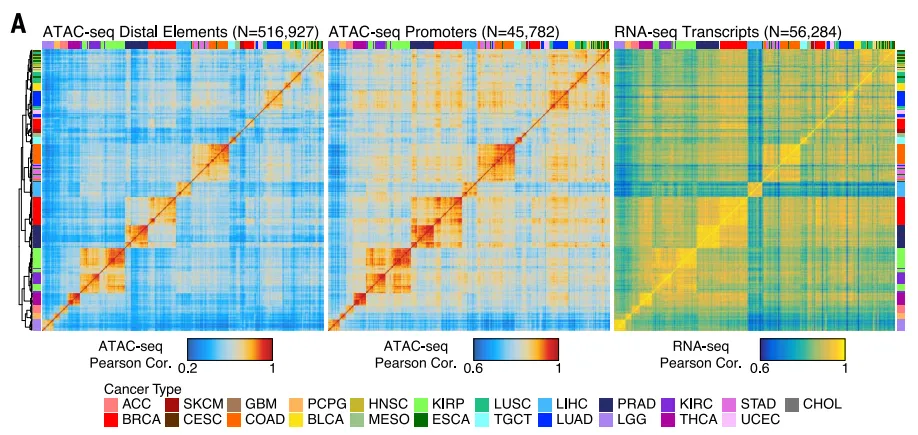
The comparison between the typing based on Distal elements and Promoters data and the typing based on RNA-seq data illustrates that ATAC-seq data can be used to distinguish different tumor types.
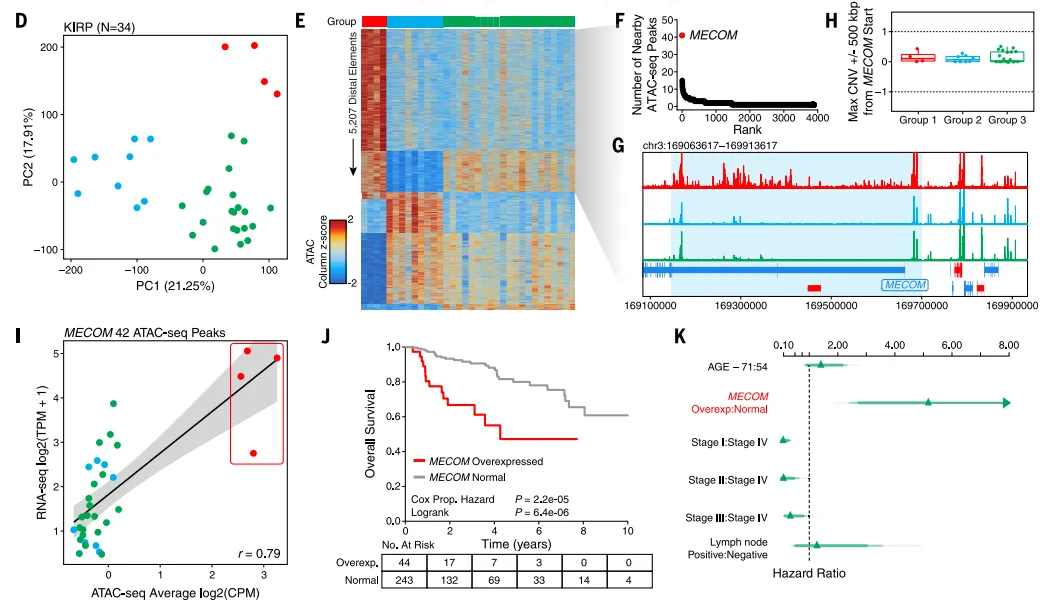
The molecular typing study of KIRP (a type of kidney cancer) using ATAC-seq data found that the red part in the figure represents a new subtype. In this subtype, the chromatin binding activity of the MECOM gene is abnormally increased, which leads to high expression of the MECOM gene. The overexpression of MECOM is significantly associated with poorer overall survival.
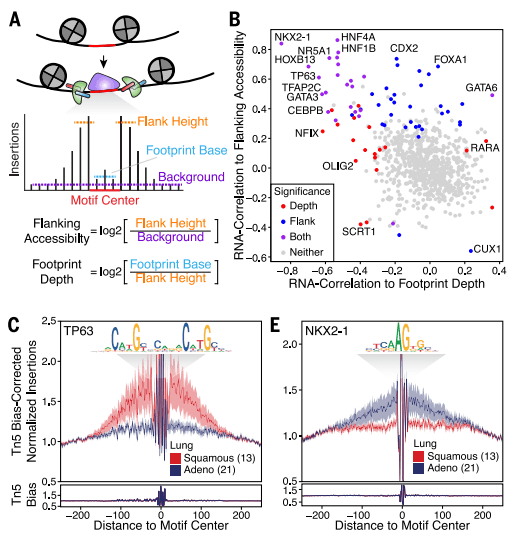
Through the search for specific peaks in different types, it was found that these specific peaks are mainly transcription factor binding sites and are closely related to methylation. Moreover, specific transcription factors for different tumor types were also discovered.
By using ATAC-seq data and RNA-seq data to calculate the correlation between chromatin accessibility and gene expression, a total of 8,552 peaks corresponding to the expression regulation of protein-coding genes were predicted. The specific peaks in different tumor types are different.
Reference
[1] Grandi F C, Modi H, Kampman L, et al. Chromatin accessibility profiling by ATAC-seq[J]. Nature protocols, 2022, 17(6): 1518-1552.
[2] M. Ryan Corces et al.,The chromatin accessibility landscape of primary human cancers.Science362,eaav1898(2018).DOI:10.1126/science.aav1898.
