introduction
Parallel Reaction Monitoring (PRM) is an ion detection technique based on high-resolution and high-precision mass spectrometry. It can selectively detect target proteins and target peptides, thereby achieving relative or absolute quantification of target proteins and peptides. PRM can complement traditional protein verification techniques (such as Western Blot/ELISA), making protein verification simpler and offering more options. It is typically applicable to the verification of high-throughput proteomic quantitative data and the quantitative detection of biomarker protein molecules in clinical samples.
advantages
✔ Quantification based on unique peptides leads to more reliable results.
✔ Dozens of proteins can be detected at one time.
✔ The cycle is clear and the risks are controllable.
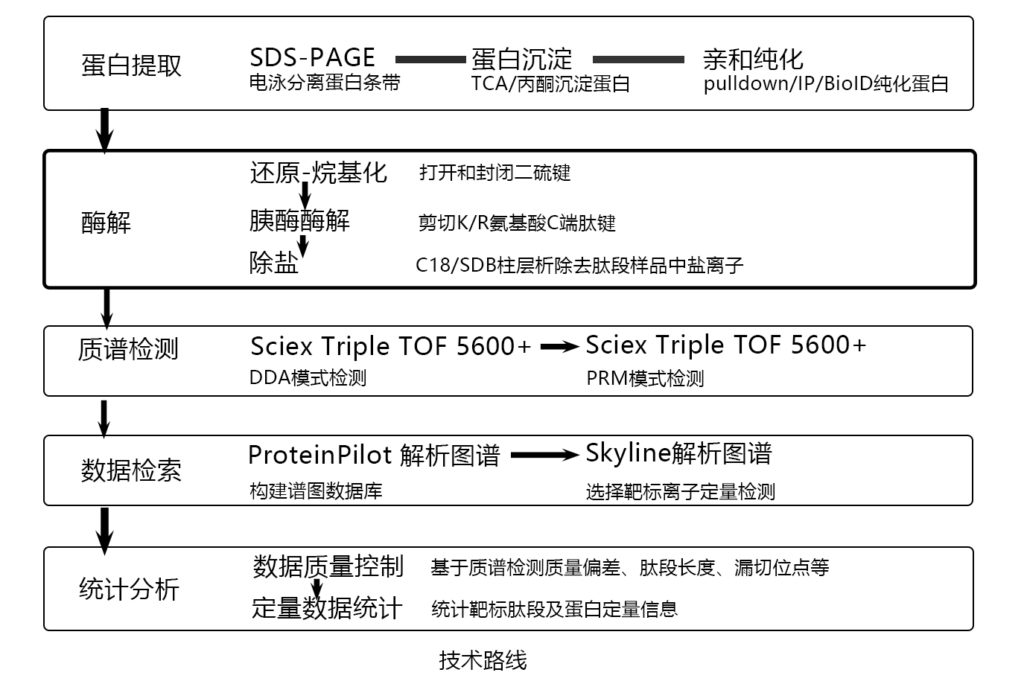
workflow
.png)
research Cases
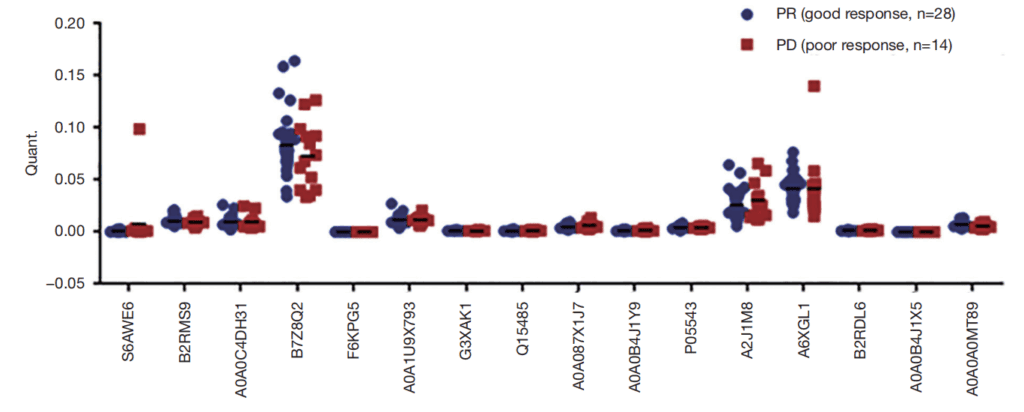
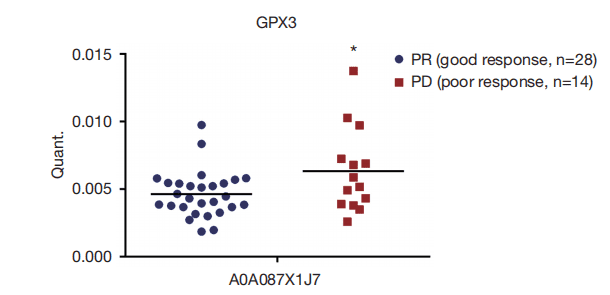
▶ Identify serum biomarkers through data-independent acquisition (DIA) mass spectrometry analysis and parallel reaction monitoring (PRM) verification to predict the efficacy of pemetrexed/platinum-based chemotherapy in patients with advanced lung adenocarcinoma (Translational Lung Cancer Research, IF = 6.496).
Pemetrexed/platinum-based chemotherapy is the standard chemotherapy regimen for patients with lung adenocarcinoma, but the efficacy varies greatly. To discover new serum biomarkers for predicting the efficacy of pemetrexed/platinum-based chemotherapy, the authors conducted data-independent acquisition (DIA) proteomics detection on serum samples from 20 patients with advanced lung adenocarcinoma who received pemetrexed/platinum-based chemotherapy. A total of 23 differentially expressed proteins were obtained by comparing the group with good chemotherapy response (PR) and the group with poor chemotherapy response (PD). Among them, the ROC AUC values of 7 proteins for predicting treatment response were higher than 0.8. To further verify the results of DIA proteomics, the authors used parallel reaction monitoring (PRM) to detect 16 candidate serum biomarkers in the study cohort of these 20 patients and a new cohort containing 22 patients with advanced lung adenocarcinoma (16 PR and 6 PD). The PRM verification showed good consistency with DIA proteomics, and 10 differentially expressed proteins exhibited similar up-regulation or down-regulation. Overall, through DIA proteomics and PRM verification, the researchers have found potential protein markers that can better reflect the treatment response of advanced lung adenocarcinoma.