introduction
LC-MS/MS Protein identification uses a high-resolution liquid chromatography-mass spectrometry (LC-MS/MS) combined system to conduct mass spectrometry detection and analysis on the target bands, and reports and gives feedback in the form of a protein identification list sorted according to protein abundance. Its basic principle is to utilize the ion peak distribution of the tandem mass spectrometry obtained by the mass spectrometer to identify the peptide segments in the samples and then identify the types of proteins.
advantages
✔ High detection sensitivity.
✔ High accuracy.
✔ The protein identification list is sorted according to the reference values of protein abundance, which is beneficial for screening.
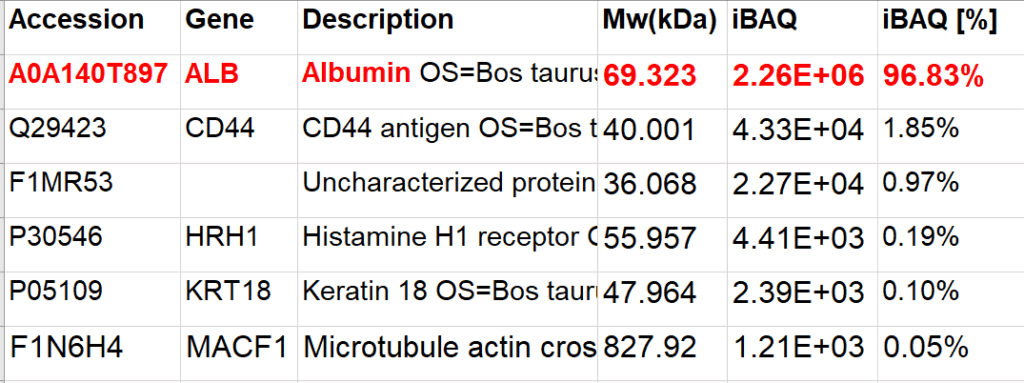
Demo Data
Research Cases
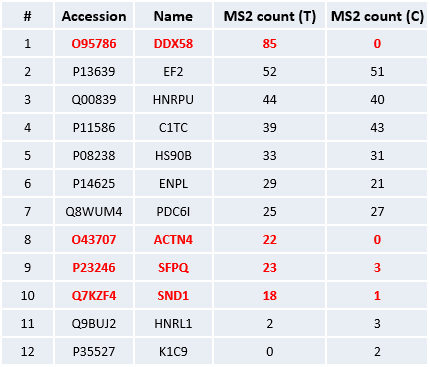
Identify the proteins that interact with the target protein DDX58. To achieve this goal, two cell lines were selected. One was the 293 cell line stably expressing the HA-tagged DDX58 protein, named the experimental group T; the other was the conventional 293 cell line, named the control group C. Immunoprecipitation was carried out on the experimental group T and the control group C using the HA antibody respectively. Then the protein eluates were collected. After enzymatic digestion in solution, mass spectrometry analysis was performed to exclude non-specific binding proteins, and finally the proteins that interact with DDX58 were screened out.
The proteins identified in the experimental group T and the control group C can be divided into two major categories: one category consists of proteins whose amounts are higher in group T than in group C, and the other category includes proteins whose amounts are equivalent or higher in group C. According to the purpose of the experiment, the proteins of the first category are the ones that need attention and are regarded as candidate proteins for interaction. Part of the identification results are shown in the table below. The target protein DDX58 was only detected in the experimental group T, while ACTN4, SFPQ, and SND1 may be the interacting proteins of DDX58. Based on the principle that the higher the protein abundance is, the greater the number of acquired tandem mass spectra will be, we adopted the spectral counting method to conduct a relative comparison of protein contents, and the results are shown in the table below.