Advantages
Is the RBP related to my research?
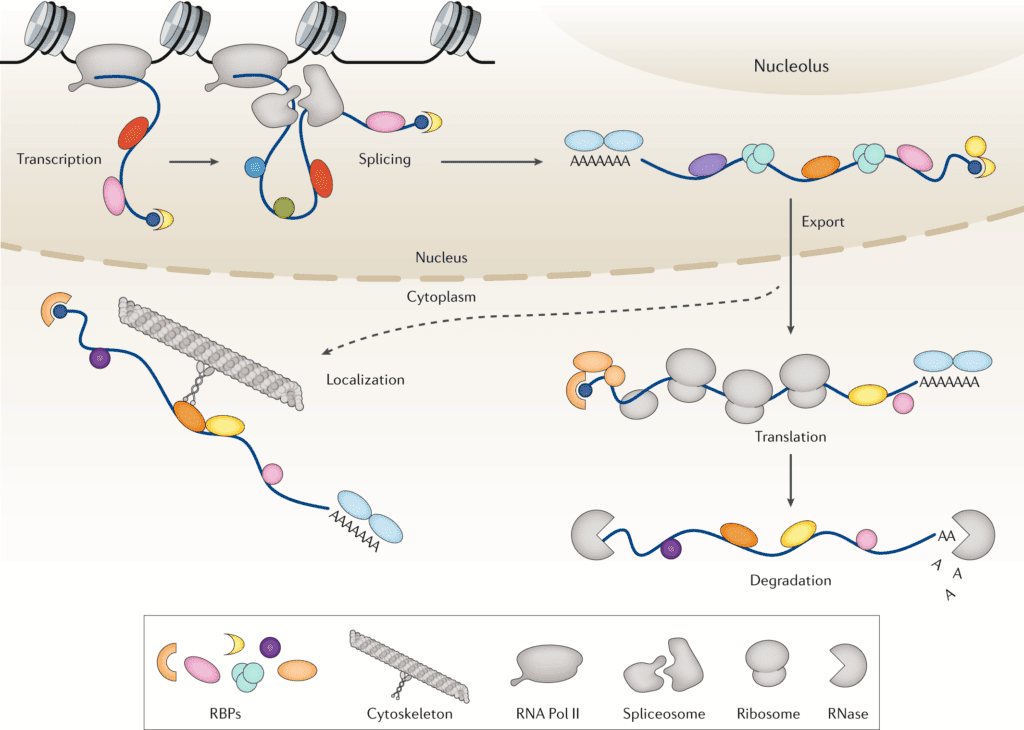
RNA binding proteins (RBPs) are a class of proteins that can specifically bind to RNA molecules. They recognize RNA targets through classic RNA binding domains (such as the RNA recognition motif RRM, DEAD-box helicase domain) or non-classic domains (such as intrinsically disordered regions). The human genome encodes more than 2,000 kinds of RBPs, and their functions run through the entire life cycle of RNA.
RNA binding proteins can bind and interact with non-coding RNAs such as circRNA, lncRNA, and miRNA. Meanwhile, they are closely related to many current hotspots, including tumor immunity, T cell immunotherapy, m6A modification, single-cell sequencing, alternative splicing, CRISPR-Cas9, etc., and there have been significant findings, which are well worth our in-depth research.
It can be said that without RBPs, RNA cannot play any physiological role.
What is the RBP research data package?
Data packages for RBP silencing/overexpression.
Through literature retrieval, database analysis and other methods, RNA binding proteins (RBPs) related to disease research are identified. According to the specific functions of RBPs, the impact of RBP silencing/overexpression on gene expression patterns, expression levels, functional pathways and alternative splicing events is studied at the overall level by using transcriptome sequencing technology (RNA-seq), so as to provide comprehensive transcriptome information. Cell function experiments can also be selected, such as cell proliferation, apoptosis, invasion, metastasis and function experiments related to specific diseases.
The iRIP data package of RBPs
The iRIP-seq technology can identify the RNAs that bind directly and indirectly to specific RNA binding proteins (RBPs), obtain information such as binding motifs, binding intensities and binding targets, which can provide more solid and direct evidence for the analysis results of silencing/overexpression products and study the functions and mechanism of action of RBPs at the overall level.
Research

Title:Insulin-like growth factor 2 mRNA-binding protein 2-regulatedalternative splicing of nuclear factor 1 C-type causes excessive granulosacell proliferation in polycystic ovary syndrome
Journal:Cell Proliferation
Publication Date:2022
Objective: Polycystic ovary syndrome (PCOS) is a common reproductive endocrine disorder. Insulin-like growth factor 2 mRNA binding protein 2 (IGF2BP2), as a target gene of HMGA2, can promote the proliferation of granulosa cells (GCs). However, it remains unclear whether IGF2BP2 participates in the pathogenesis of PCOS as an RNA binding protein (RBP). In this study, the authors aimed to clarify the transcripts that interact with IGF2BP2, the whole transcriptome, and alternative splicing in GCs, thereby revealing the potential mechanisms underlying the pathogenesis of PCOS.
Materials and Methods: Quantitative reverse transcription PCR (RT-qPCR) and Western Blot were used to detect the expression of IGF2BP2 in GCs of PCOS patients. RNA immunoprecipitation sequencing (RIP-seq) and RNA sequencing (RNA-seq) were employed to capture the transcripts that interact with IGF2BP2, the whole transcriptome, and alternative splicing. KGN cells transfected with overexpressing IGF2BP2 plasmids and nuclear factor 1 type C (NFIC) siRNA were detected by using CCK-8, EdU and TUNEL assays.
Results: IGF2BP2 was highly expressed in GCs of PCOS patients. As an RBP, IGF2BP2 preferentially bound to the 3′ and 5’UTR regions of mRNA, and the binding motifs were GGAC and the newly discovered GAAG motif. The overexpression of IGF2BP2 altered the transcriptome profile of KGN cells. IGF2BP2 promoted cell proliferation by inhibiting the exon skipping event of NFIC and regulating alternative splicing events.
