introduction
In 1958, Crick put forward the “Central Dogma”, which elaborated on the information flow among DNA, RNA and proteins. Genes are transcribed into precursor RNA, and through alternative splicing, different transcripts are formed, which are then translated into protein isoforms. Alternative splicing enables genes to produce multiple isoforms with different structures and functions, and their expressions are specific. Recently, “Nature Reviews Drug Discovery” has explored the role of protein isoforms in drug discovery and proposed to utilize isoforms to enhance drug specificity and efficacy. Therefore, studying the regulatory mechanisms of alternative splicing of precursor RNA and the translation mechanisms of transcripts is crucial for the development of treatments centered around protein isoforms.
The Poly(A) tail is a key structural element of mRNA and is essential for the stability and translation process of mRNA. However, we often overlook the epigenetic regulatory mechanisms behind it. The progress of sequencing technologies enables us to comprehensively and in-depth analyze the length and composition of the Poly(A) tail, revealing its abundant regulatory information and providing new perspectives and possibilities for the regulation of gene expression.
The third-generation sequencing technologies represented by Nanopore are insensitive to homopolymeric sequences, enabling them to perform precise Poly(A) tail sequencing. The Nanopore platform is particularly outstanding. It can not only obtain the length of the PolyA tail through DRS sequencing, but also some researchers have developed techniques that can be adapted to the ONT platform to sequence the length of the Poly(A) tail, such as FLEP-seq, FLEP-seq2 and Nano3P-seq, etc. Currently, the latest official long-read RNA library construction kit of Nanopore can also sequence the length of the Poly(A) tail. The principle of library construction is different from that of ONT-IrRNA-seq (Oxford Nanopore Technologies long-read RNA sequencing). To distinguish it from the ONT-IrRNA-seq sequencing technology, we call the new library construction and sequencing technology: ONT-PAIrRNA-seq (Oxford Nanopore Technologies Poly(A) inclusive long-read RNA sequencing). Whether through cDNA sequencing or DRS sequencing, the Nanopore sequencing technology can reveal the non-A residues within the Poly(A) tail, and this characteristic provides strong support for its application in the field of RNA research.
advantages
✔ Improve gene annotation: Discover new transcripts and genes, and obtain a comprehensive transcriptome map.
✔ Differential analysis: Differentially expressed genes (DEG), differentially expressed transcripts (DET), differentially used transcripts (DTU).
✔ Gene structure analysis: Accurately analyze alternative splicing (AS), polyadenylation sites (APA), the length of 3’UTR, and the length of polyA.
✔ Transcript resolution: Analyze the regulatory mechanisms and functions of gene expression at the transcript level.
✔ Absolute quantification: UMI sequences can accurately quantify transcripts.
workflow
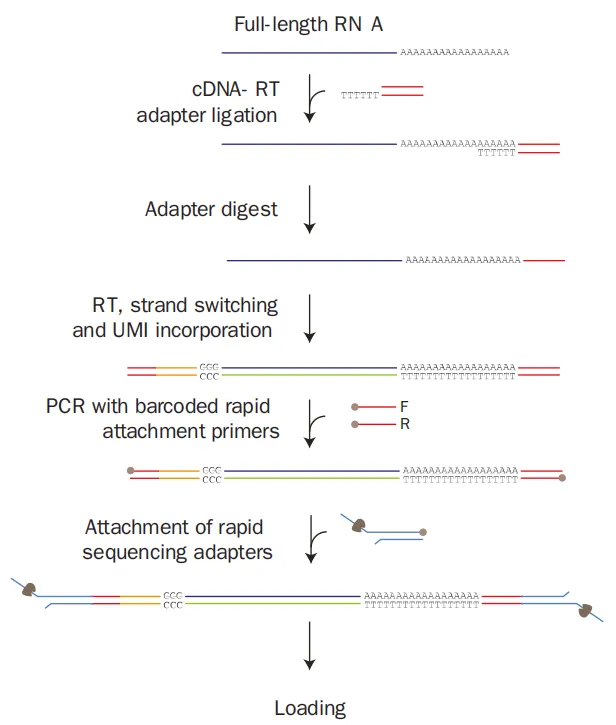
Firstly, RNA containing a polyA tail is captured by olig dT magnetic beads. Then, a double-stranded primer with a fixed sequence and a 6-T-base overhang is ligated to the last A base at the 3’ end of the RNA using T4 DNA ligase. After that, the unligated strand of the double-stranded primer is digested. Subsequently, an RT primer is added for reverse transcription, and the TSO + UMI sequence is added to the 3’ end of the cDNA through strand displacement. Next, primer PCR amplification is carried out to form double-stranded cDNA. Finally, the ends of the double-stranded cDNA are blunted and ligated with ONT sequencing adapters to form a sequencing library.
research cases
Case 1
Nano3P-seq: Transcriptome-wide Analysis of Gene Expression and Tail Dynamics Using End-capture Nanopore cDNA Sequencing
Journal: Nature Methods
Impact Factor (IF): 36.1
Technical Method: Nanopore 3′ end-capture sequencing
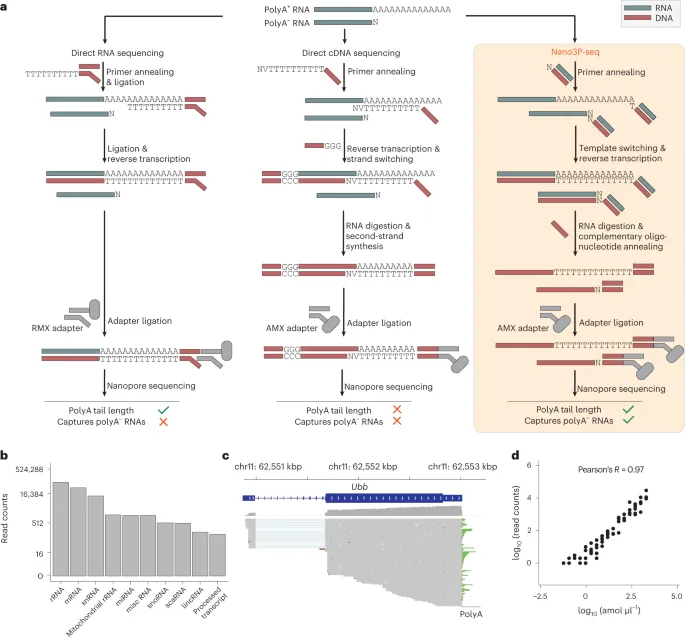
RNA polyadenylation plays a central role in RNA maturation, fate, and stability. Variations in the length of the polyA tail affect the translation efficiency and stability of mRNA. The authors developed Nano3P-seq, which utilizes nanopore cDNA sequencing to quantify RNA abundance, tail composition, and length dynamics. Nano3P-seq can start sequencing from the 3′ end of RNA molecules without the need for PCR amplification or RNA adapter ligation. This method enables the quantitative estimation of RNA abundance and tail length and can capture multiple RNA biotypes. In mice and zebrafish, it was found that 16S mitochondrial rRNA also has a polyadenylate tail. The length of the mRNA tail is dynamically regulated during vertebrate embryogenesis and is associated with mRNA decay. Nano3P-seq can capture non-A bases within polyA tails of different lengths and reveal their distribution during vertebrate embryogenesis. Overall, Nano3P-seq is a simple and robust method that accurately estimates transcriptional levels, tail length, and heterogeneity in tail composition with minimal bias in library preparation.
Case 2
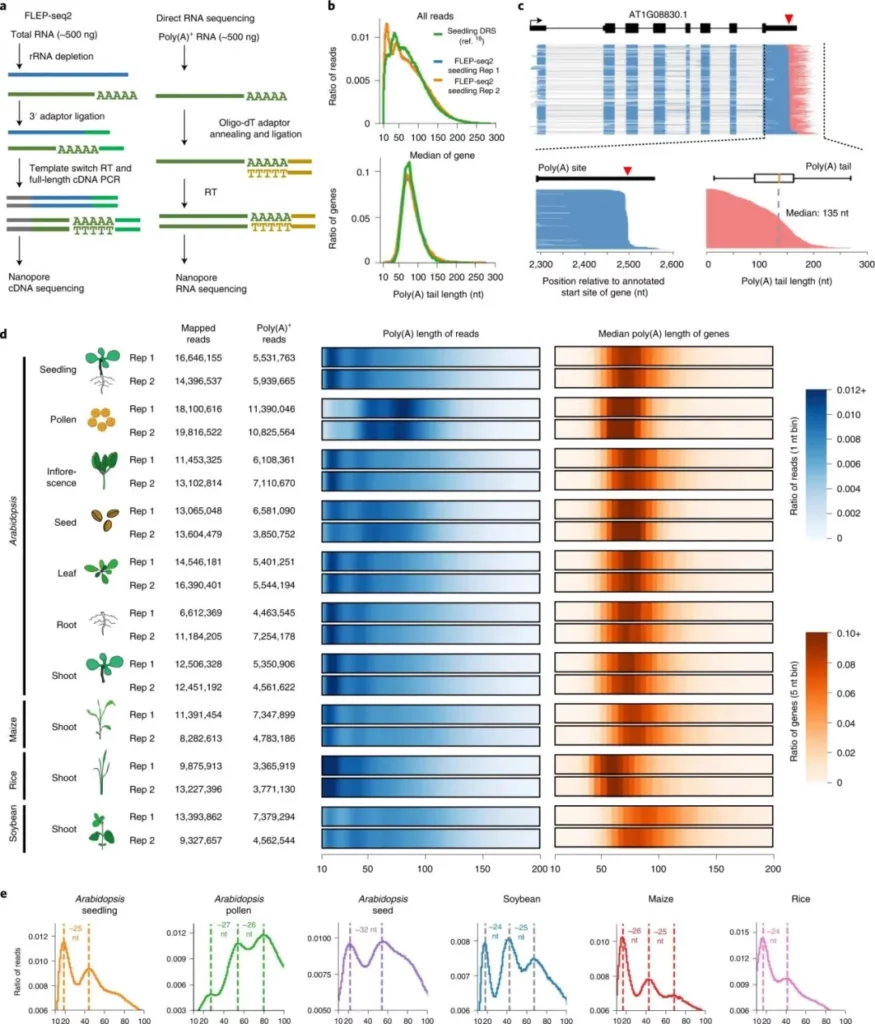
An atlas of plant full-length RNA reveals tissue-specific and monocots–dicots conserved regulation of poly(A) tail length.
Journal: Nature Plants
Impact Factor (IF): 15.8
Samples: Seven different tissues of Arabidopsis thaliana
Technical Method: FLEP-seq2
参考文献
【1】 Kjer-Hansen, P., Phan, T.G. & Weatheritt, R.J. Protein isoform-centric therapeutics: expanding targets and increasing specificity. Nat Rev Drug Discov (2024).
【2】Jia, J., Lu, W., Liu, B. et al. An atlas of plant full-length RNA reveals tissue-specific and monocots–dicots conserved regulation of poly(A) tail length. Nat. Plants 8, 1118–1126 (2022).
