RXBio Translates Sequence to Science and Industry
Tel: 027-87050299Email: sales@rxbio.cc
- Home
- Single-cell Sequencing
- Spatial Transcriptomics
- Third-generation Sequencing
- Omics Technologies
- Bioinformatics
- Experimental Platform
- About US
中文
RXBio Translates Sequence to Science and Industry
In eukaryotic cells, the 3′ ends of most protein-coding genes and long non-coding RNAs undergo cleavage and polyadenylation (polyA). Recent studies have shown that the vast majority of mRNAs have two or more polyadenylation sites (PAS). Therefore, alternative polyadenylation (APA, referring to genes with multiple PAS) is an important post-transcriptional regulatory mechanism in eukaryotes. It is widely present in all eukaryotes and is a major mechanism for gene regulation.
APA events can not only lead to the generation of multiple transcriptional isoforms with different lengths and sequence compositions of the 3′ untranslated region (3’UTR) for coding genes, expanding proteomic and functional diversity, but also play an important role in gene regulation. As a key molecular mechanism, APA participates in multiple gene regulation steps such as mRNA maturation, mRNA stability, cellular RNA decay, and protein diversification. APA is often dysregulated in cancer, resulting in changes in the expression of oncogenes and tumor suppressor genes. In normal cells, oncogenes usually use distal PAS to form long 3’UTR subtypes, while tumor cells tend to use proximal PAS to form short 3’UTR subtypes. The shortening of the 3’UTR may significantly reduce the inhibitory effect of microRNAs, make the mRNA more stable, significantly improve the translation efficiency, and increase the protein expression level of related genes.
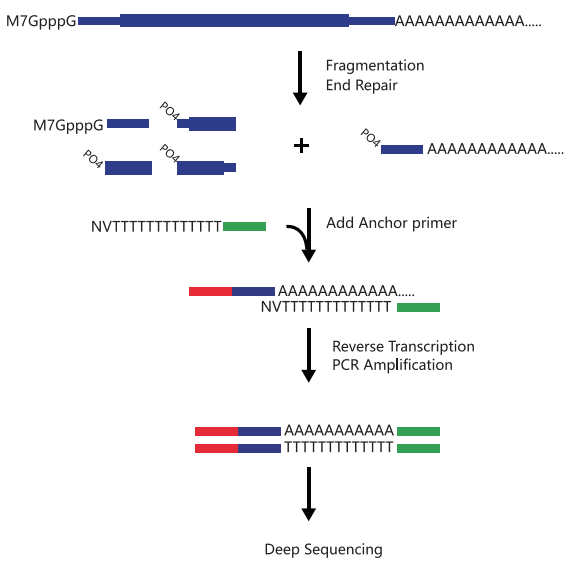
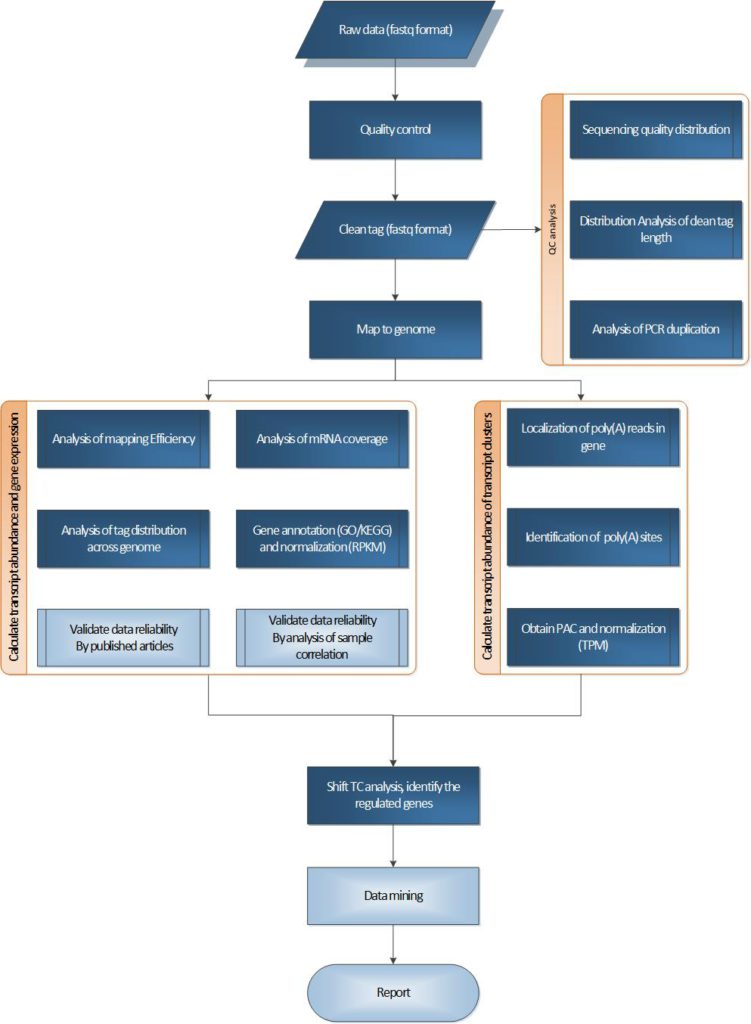
PAS-seq is a technical method for detecting variable polyadenylation sites. Alternative polyadenylation (APA) is an important gene expression regulatory mechanism in eukaryotes. Through PAS-seq and subsequent informatics analysis, we can obtain transcription termination sites (TTS) within the transcriptome, different 3’UTR analyses, and differential expression analyses.
✔ Direct and convenient
✔ Easy to achieve
✔ Own analysis process
✔ Personalized service support
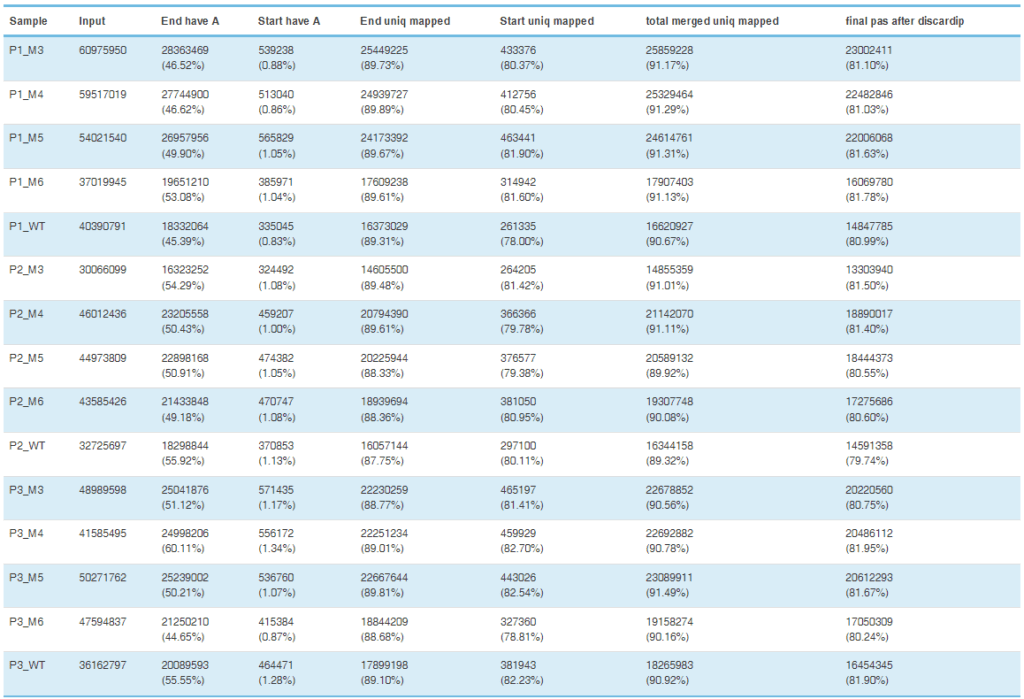
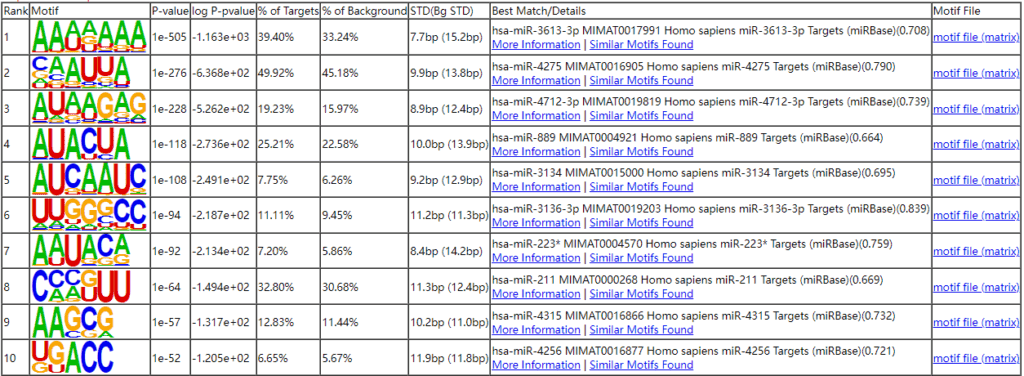
The important regulatory role of alternative polyadenylation (APA) in suppressing cervical cancer.
Journal: Oncogene, with an Impact Factor (IF) of 9.867.
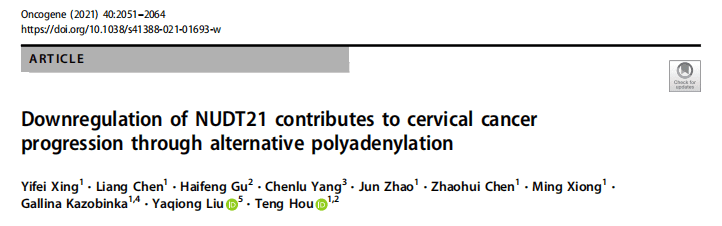
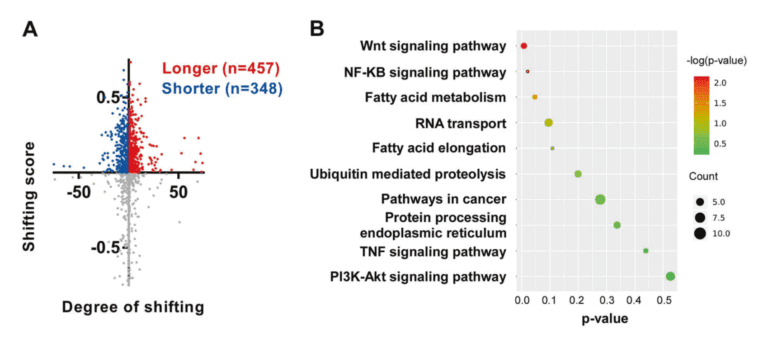
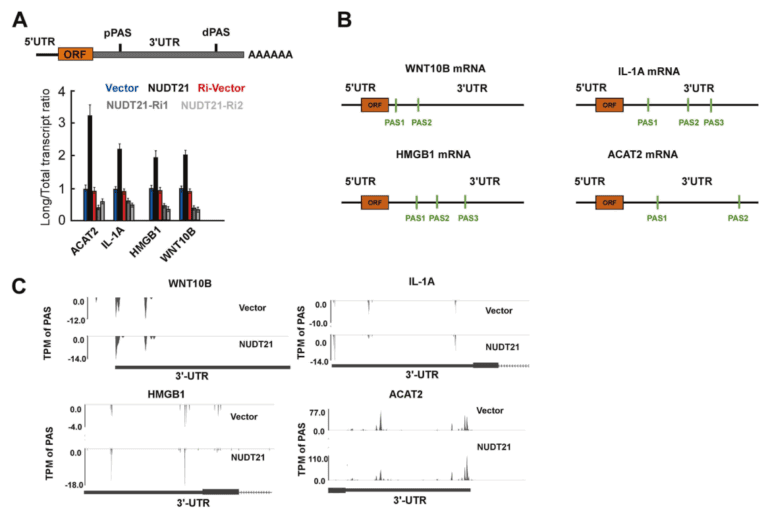
Utilize our company’s unique PAS-seq technology to sequence the 3′ untranslated region (3’UTR) of polyadenylated RNA within the whole genome of cervical cancer, map the alternative polyadenylation (APA) events in response to the overexpression and knockdown of NUDT21, and then conduct functional annotation analysis. In the case of NUDT21 overexpression, it was found that the 3’UTRs of 457 transcripts were significantly elongated. Meanwhile, the 3’UTRs of 348 transcripts were significantly shortened. Through functional annotation analysis, it was discovered that the targets of NUDT21 are involved in the roles of fatty acid (FA) metabolism regulators and the Wnt and NF-κB signaling pathways. From the genes whose APA is regulated by NUDT21 mentioned above, the authors further confirmed that IL-1A, WNT10B, HMGB1, and ACAT2 are involved in the metastasis genes of cervical cancer. NUDT21 can regulate miRNA-mediated translation inhibition in CxCa cells.